In this tutorial, we will learn the fundamentals of imitation learning by playing a video game. We will teach an agent how to play Flappy Bird, one of the most popular video games in the world way back in 2014.
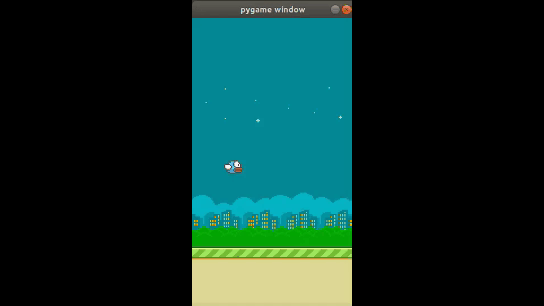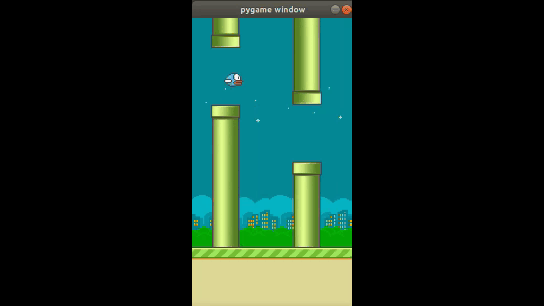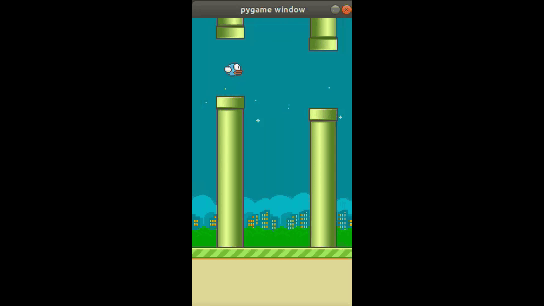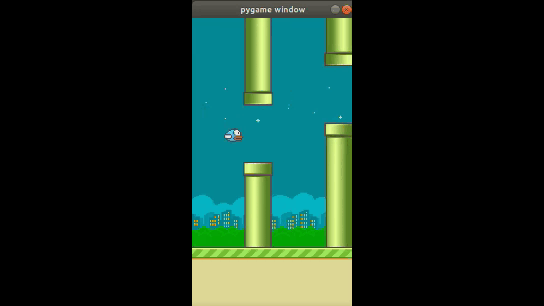
The objective of the game is to get a bird to move through a sequence of pipes without touching them. If the bird touches them, he falls out of the sky.
Table of Contents
- You Will Need
- What is Imitation Learning?
- Directions
- –Install Flappy Bird
- –Launch Flappy Bird
- –Implement the Dataset Aggregation Algorithm (DAgger)
- References
You Will Need
- Python 3.6 (or higher)
- Ubuntu Linux (If you’re on Windows or MacOS, you can download a virtual machine and install Ubuntu. Don’t be scared if you’ve never used Ubuntu Linux before. It is a great operating system, and you’ll get comfortable with it in no time.)
- An account with GitHub.
What is Imitation Learning?
In order to understand what imitation learning is, it is first helpful to understand what reinforcement learning is all about.
Reinforcement learning is a machine learning method that involves training a software “agent” (i.e. algorithm) to learn through trial and error. The agent is rewarded by the environment when it does something good and punished when it does something bad. “Good” and “bad” behavior in this case are defined beforehand by the programmer.
The idea behind reinforcement learning is that the agent learns over time the best actions it must take in different situations (i.e. states of the environment) in order to maximize its rewards and minimize its punishment. That stuff in bold in the previous sentence is formally known as the agent’s optimal policy.
Check out my reinforcement learning post for an example of how reinforcement learning works.
One of the problems with reinforcement learning is that it takes a long time for an agent to learn through trial and error. Knowing the optimal action to take in any given state of the environment means that the agent has to explore a lot of different random actions in different states of the environment in order to fully learn.
You can imagine how infeasible this gets as the environment gets more complex. Imagine a robot learning how to make a breakfast of bacon and eggs. Assume the robot is in your kitchen and has no clue what bacon and eggs are. It has no clue how to use a stove. It has no clue how to use a spatula. It doesn’t even know how to use a frying pan.

The only thing the robot has is the feedback it receives when it takes an action that is in accordance with the goal of getting it to make bacon and eggs.
You could imagine that it could take decades for a robot to learn how to cook bacon and eggs this way, through trial and error. As humans, we don’t have that much time, so we need to use some other machine learning method that is more efficient.
And this brings us to imitation learning. Imitation learning helps address the shortcomings of reinforcement learning. It is also known as learning from demonstrations.
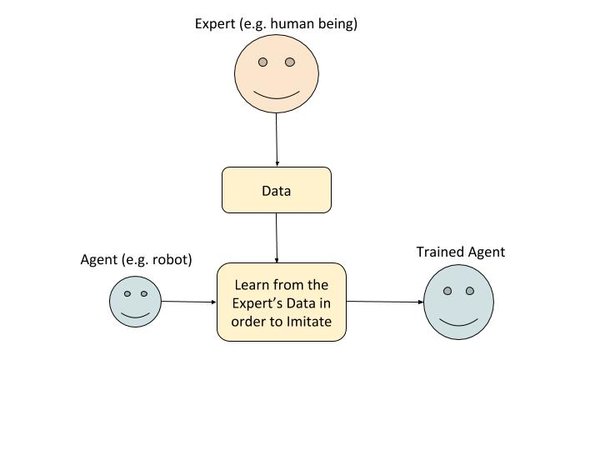
In imitation learning, during the training phase, the agent first observes the actions of an expert (e.g. a human). The agent keeps a record of what actions the expert took in different states of the environment. The agent uses this data to create a policy that attempts to mimic the expert.
Babies and little kids learn this way. They imitate the actions of their parents. For example, I remember learning how to make my bed by watching my mom and then trying to imitate what she did.
Now, at runtime, the environment won’t be exactly similar as it was when the agent was in the training phase observing the expert. The agent will try to approximate the best action to take in any given state based on what it learned while watching the expert during the training phase.
Imitation learning is a powerful technique that works well when:
- The number of states of the environment is large (e.g. in a kitchen, where you can have near infinite ways a kitchen can be arranged)
- The rewards are sparse (e.g. there are only a few actions a robot can take that will bring him closer to the goal of making a bacon and egg breakfast)
- An expert is available.
Going back to our bacon and eggs example, a robotic agent doing imitation learning would observe a human making bacon and eggs from scratch. The robot would then try to imitate that process.
In the case of reinforcement learning, the robotic agent will take random actions and will receive a reward every time it takes an action that is consistent with the goal of cooking bacon and eggs.
For example, if the robot cracks an egg into a frying pan, it would receive a reward. If the robot cracks an egg on the floor, it would not receive a reward.
After a considerable amount of time (maybe years!), the robot would finally be smart enough (based on performance metrics established by the programmer) to know how to cook bacon and eggs.
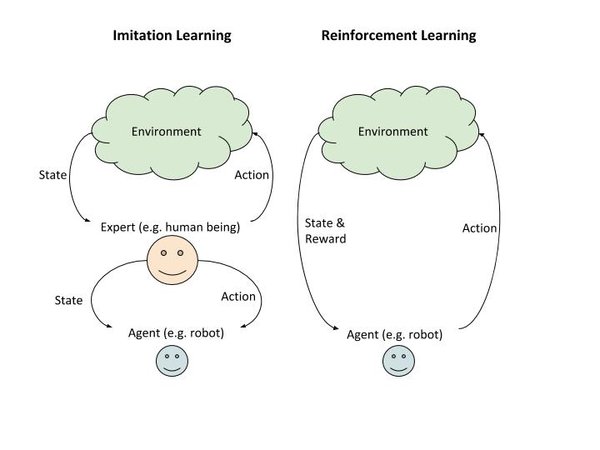
So you see that the runtime behavior of imitation learning and reinforcement learning are the same. The difference is in how the agent learns during the training phase.
In reinforcement learning, the agent learns through trial and error by receiving varying rewards from the environment in response to its action.
In imitation learning, the agent learns by watching an expert. It observes the action the expert took in various states.
Because there is no explicit reward in the case of imitation learning, the agent only becomes as good as the teacher (but never better). Thus, if you have a bad expert, you will have a bad agent.
The agent doesn’t know the reward (consequences) of taking a specific action in a certain state. The graphic below further illustrates the difference between reinforcement learning and imitation learning.
So with that background, let’s dive into implementing an imitation learning algorithm to teach an agent to play Flappy Bird.
Directions
Install Flappy Bird
You can find the instructions for installing FlappyBird at this link, but let’s run through the process as it can be a bit tricky. Go slow, so you make sure you get all the necessary libraries.
Open up a terminal window.
Install all the dependencies for Python 3.X listed here. I’ll copy and paste all that below, but if you have any issues, check out the link. You can copy and paste the command below into your terminal.
sudo apt-get install git python3-dev python3-setuptools python3-numpy python3-opengl \ libsdl-image1.2-dev libsdl-mixer1.2-dev libsdl-ttf2.0-dev libsmpeg-dev \ libsdl1.2-dev libportmidi-dev libswscale-dev libavformat-dev libavcodec-dev \ libtiff5-dev libx11-6 libx11-dev fluid-soundfont-gm timgm6mb-soundfont \ xfonts-base xfonts-100dpi xfonts-75dpi xfonts-cyrillic fontconfig fonts-freefont-ttf libfreetype6-dev
You don’t have to know what each library does, but in case you’re interested, you can take a look at the Ubuntu packages page to get a complete description.
Make sure you have pip installed.
Now you need to install pygame.
pip3 install pygame
We’re not done yet with all the installations. Let’s install the PyGame Learning Environment (PLE) now by copying the entire repository to our computer. It already includes Flappy Bird.
git clone https://github.com/ntasfi/PyGame-Learning-Environment
You will be asked to login to GitHub. Type in your username and password.
Type the following command to see if it installed properly. You should see the PyGame-Learning-Environment folder.
ls
Now let’s switch to that folder.
cd PyGame-Learning-Environment
Complete the install, by typing the following command (be sure to include the period).
sudo pip3 install -e .
Now Flappy Bird is all set up.
Test to see if everything is working, type the following command to run the Aliens program
python3 -m pygame.examples.aliens
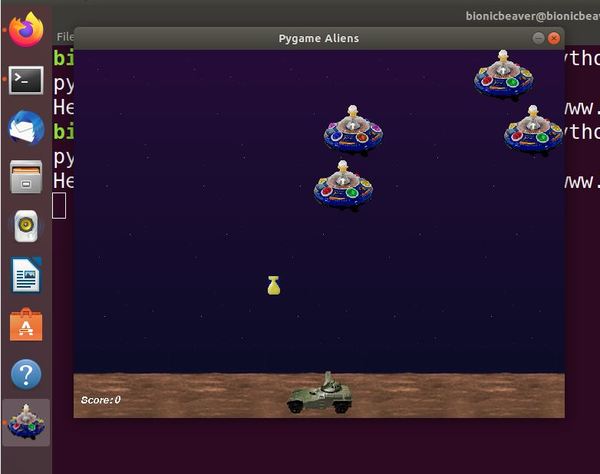
Launch Flappy Bird
Let’s write a Python script in order to use the game environment.
Open a new terminal window.
Go to the PyGame-Learning-Environment directory.
cd PyGame-Learning-Environment
We will create the script flappybird1.py. Open the text editor to create a new file.
gedit flappybird1.py
Here is the full code. You can find a full explanation of the methods at the PyGameLearning Environment website. The function getScreenRGB retrieves the state of the environment in the form of red/green/blue image pixels:
# Import Flappy Bird from games library in
# Python Learning Environment (PLE)
from ple.games.flappybird import FlappyBird
# Import PyGame Learning Environment
from ple import PLE
# Create a game instance
game = FlappyBird()
# Pass the game instance to the PLE
p = PLE(game, fps=30, display_screen=True, force_fps=False)
# Initialize the environment
p.init()
actions = p.getActionSet()
print(actions) # 119 to flap wings, or None to do nothing
action_dict = {0: actions[1], 1:actions[0]}
reward = 0.0
for i in range(10000):
if p.game_over():
p.reset_game()
state = p.getScreenRGB()
action = 1
reward = p.act(action_dict[action])
Now, run the program using the following command:
python3 flappybird1.py
Here is what you should see:
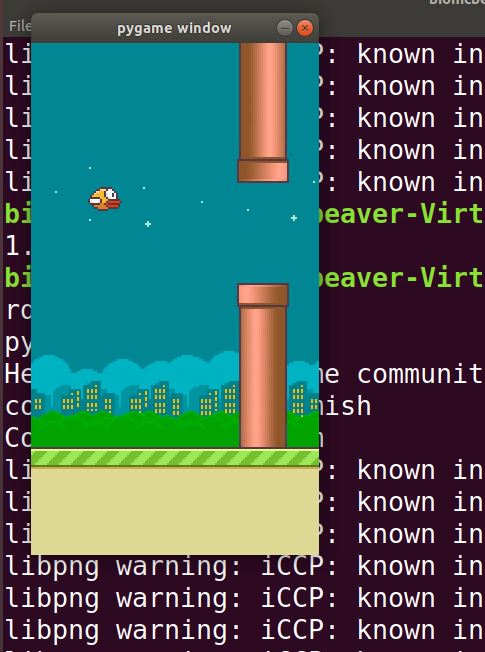
Here is flappybird2.py. The agent takes a random action at each iteration of the game.
# Import Flappy Bird from games library in
# Python Learning Environment (PLE)
from ple.games.flappybird import FlappyBird
# Import PyGame Learning Environment
from ple import PLE
# Import Numpy
import numpy as np
class NaiveAgent():
"""
This is a naive agent that just picks actions at random.
"""
def __init__(self,actions):
self.actions = actions
def pickAction(self, reward, obs):
return self.actions[np.random.randint(0, len(self.actions))]
# Create a game instance
game = FlappyBird()
# Pass the game instance to the PLE
p = PLE(game)
# Create the agent
agent = NaiveAgent(p.getActionSet())
# Initialize the environment
p.init()
actions = p.getActionSet()
action_dict = {0: actions[1], 1:actions[0]}
reward = 0.0
for f in range(15000):
# If the game is over
if p.game_over():
p.reset_game()
action = agent.pickAction(reward, p.getScreenRGB())
reward = p.act(action)
if f > 1000:
p.display_screen = True
p.force_fps = False # Slow screen
Implement the Dataset Aggregation Algorithm (DAgger)
How the DAgger Algorithm Works
In this section, we will implement the Dataset Aggregation Algorithm (DAgger) and apply it to FlappyBird.
The DAgger algorithm is a type of imitation learning algorithm that is intended to help address the problem of the agent “memorizing” the actions of the expert in different states. It is time-consuming to watch the expert do every combination of possible states of the environment, so we need the expert to give the agent some guidance on what to do in the event the agent makes errors.
In the first phase, we first let the expert take action in the environment and keep track of what action the expert took in a variety of states of the environment. This phase creates an initial “policy”, a collection of state-expert action pairs.
Then for the second phase, we add a twist to the situation. The expert continues to act, and we record the expert’s actions. However, the environment doesn’t change according to the expert’s actions. Rather it changes in response to the policy from phase 1. The environment is “ignoring” the actions of the expert.
In the meantime, a new policy is created that maps the state to the expert’s actions.
You might now wonder, which policy would the agent choose? The policy from phase 1 or phase 2. One method is to pick one at random. Another method (and the one often preferred in practice) is to hold out a subset of states of the environment and test which one performs the best.
You can find the Dagger paper here to learn more.
Install TensorFlow
We now need to install TensorFlow, the popular machine learning library.
Open a new terminal window, and type the following command (wait a bit as it will take some time to download).
pip3 install tensorflow
Run FlappyBird
Now we need to run FlappyBird with the imitation learning algorithm. Since the game has no real expert, we have to have our algorithm load a special file (included in the GitHub folder that we will clone in a second) that has the expert’s pre-prepared policy.
Clone the code at this GitHub page here into the file.
git clone https://github.com/PacktPublishing/Reinforcement-Learning-Algorithms-with-Python.git
To run the code, migrate to the “Chapter10” file. You want to find the file called Dagger.py.
gedit Dagger.py
To see FlappyBird in action, change this line:
env = PLE(env, fps=30, display_screen=False)
To this:
env = PLE(env, fps=30, display_screen=True, force_fps=False)
To run the code, type the following command:
python3 Dagger.py
Eventually, you should see the results of each game spit out, with the bird’s score on the far right. Notice that the agent’s score gets above 100 after only a few minutes of training.

That’s it for imitation learning. If you want to learn more, I highly suggest you check out Andrea Lonza’s book, Reinforcement Learning Algorithms with Python, that does a great job teaching imitation learning.
References
- Reinforcement Learning Algorithms with Python by Andrea Lonza
- Applying Q-Learning to Flappy Bird by Moritz Ebeling-Rump, Manfred Kao, Zachary Hervieux-Moore
- Playing FlappyBird with Deep Reinforcement Learning by Naveen Appiah and Sagar Vare