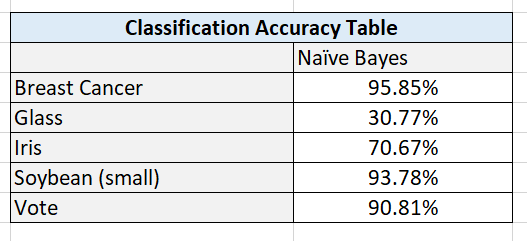
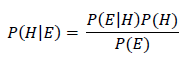In this post, I will walk you through the Winnow2 machine learning algorithm, step-by-step. We will develop the code for the algorithm from scratch using Python. We will then run the algorithm on a real-world data set, the breast cancer data set from the UCI Machine Learning Repository. Our goal is to be able to predict if a patient has breast cancer or not based on ten different attributes. Without further ado, let’s get started!
Table of Contents
- What is the Winnow2 Algorithm?
- Algorithm Steps
- Promotion and Demotion: Building the Prediction Model One Instance at a Time
- Winnow2 Example Using Real Data
- Winnow2 Implementation
- Preprocessing the Example Data Set – Breast Cancer
- Winnow2 Algorithm in Python, Coded From Scratch
- Output Statistics of Winnow2 on the Breast Cancer Data Set
What is the Winnow2 Algorithm?
The Winnow2 algorithm was invented by Nick Littlestone in the late 1980s. Winnow2 is an example of supervised learning. Supervised learning is the most common type of machine learning.
In supervised learning, you feed the machine learning algorithm data that contains a set of attributes (also known as x, predictors, inputs, features, etc.) and the corresponding classification (also known as label, output, target variable, correct answer, etc.). Each instance (also known as data point, example, etc.) helps the Learner (the prediction mathematical model we are trying to build) learn the association between the attributes and the class. The job of the Learner is to create a model that enables it to use just the values of the attributes to predict the class.
For example, consider how a supervised learning algorithm would approach the breast cancer data set that has 699 instances. Each instance is a different medical patient case seen by a doctor.

For each instance, we have:
- 9 attributes: clump thickness, uniformity of cell size, uniformity of cell shape, marginal adhesion, single epithelial cell size, bare nuclei, bland chromatin, normal nucleoli, and mitoses.
- 2 classes: malignant (breast cancer detected) or benign (breast cancer not detected)
The end goal of a classification supervised learning algorithm like Winnow2 is to develop a model that can accurately predict if a new patient has breast cancer or not based on his or her attribute values. And in order to do that, the Learner must be trained on the existing data set in order to learn the association between the 9 attributes and the class.
In Winnow2, we need to preprocess a data set so that both the attributes and the class are binary values, 0 (zero) or 1 (one). For example, in the example I presented above, the class value for each instance is either 0 (benign – breast cancer not detected) or 1 (malignant – breast cancer detected).
Algorithm Steps
The Winnow2 algorithm continuously updates as each new instance arrives (i.e. an online learning algorithm). Here is how Winnow2 works on a high level:
Step 1: Learner (prediction model) receives an instance (data point):
Since Winnow2 can only work with 0s and 1s, the attribute values need to have already been preprocessed to be either 0 or 1. For example, using the breast cancer data example, attributes like clump thickness need to first be binarized so that they are either 0 or 1. One-hot encoding is the method used in this project in order to perform this binarization.
Step 2: Learner makes a prediction of the class the instance belongs to based on the values of the attributes in the instance
For example, does the Learner believe the patient belongs to the breast cancer class based on the values of the attributes? The Learner predicts 0 if no (benign) or 1 if yes (malignant).
Step 3: Is the Prediction Correct?
The Learner is told the correct class after it makes a prediction. If the learner is correct, nothing happens. Learning (i.e. amending the mathematical model) only occurs when the Learner gets an incorrect answer.
There are two ways for the prediction to be incorrect:
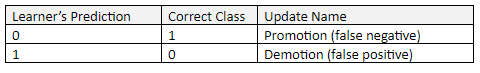
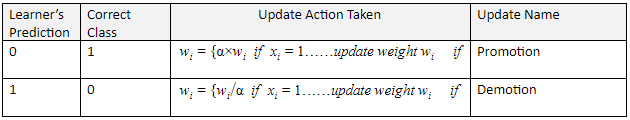
In order to “learn”, a process called “Promotion” (to be explained in the next section) takes place when the Learner incorrectly predicts 0. This situation is a “false negative” result. A false negative occurs when the Learner predicts a person does not have a disease or condition when the person actually does have it.
“Demotion” takes place when the Learner incorrectly predicts 1. This situation is a “false positive” result. A false positive (also known as “false alarm”) occurs when the Learner predicts a person has a specific disease or condition when the person actually does not have it.
Promotion and Demotion: Building the Prediction Model One Instance at a Time
To understand the promotion and demotion activities of Winnow2, we need to first examine the Learner’s mathematical model, the tool being used to predict whether an instance belongs to the benign (0) class or malignant (1) class.
When the Learner receives an instance (“Step 1” from the previous section), it runs the attributes through the following weighted sum:
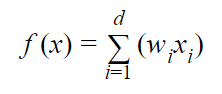
where:
- d is the total number of attributes
- wi is the weighting of the ith attribute
- xi is the value of the ith attribute in binary format
- f(x) is the weighted sum (e.g. w1x1 + w2x2 + w3x3 + …. + wdxd)
The Learner then predicts an instance’s class (e.g. 1 for malignant and 0 for benign in our breast cancer example) as follows where:
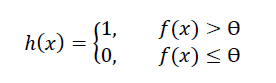
- h(x) is the predicted class
- Ɵ is a constant threshold (commonly set to 0.5)
As mentioned before, if the learner makes an incorrect prediction, either promotion or demotion occurs. In both promotion and demotion, the weights wi are modified by using a constant parameter α which is any value greater than 1 (commonly set to 2).
Initially all the weights wi for each attribute are set to 1. They are then adjusted as follows:
Winnow2 Example Using Real Data
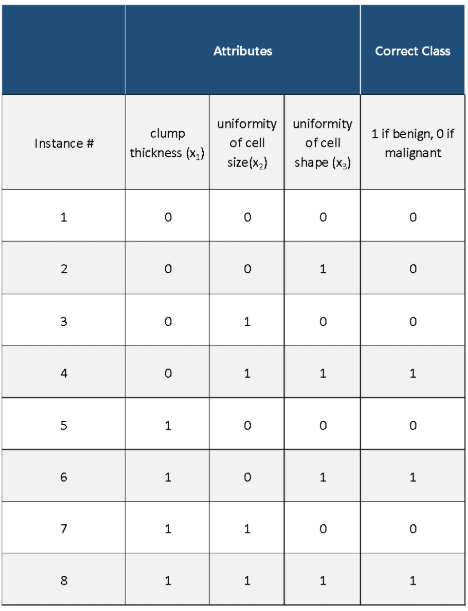
Let’s take a look at the Winnow2 algorithm using an example with real data. We have three different attributes (which have already been converted from real numbers into binary form) and a class variable (which has also been converted into binary form). Our goal is to develop a model that can use just those three attributes to predict if a medical patient has breast cancer or not.
- 3 attributes: clump thickness (x1), uniformity of cell size(x2), uniformity of cell shape (x3),
- 2 classes (labels): malignant (breast cancer detected) or benign (breast cancer not detected)
Remember:
- All weights w for each attribute are initially set to 1. This is known as the “weight vector.”
- Ɵ = 0.5. This is our threshold.
- α = 2
- d = 3 because there are 3 attributes.
Here is the original data set with the attributes (inputs) and the class (output):
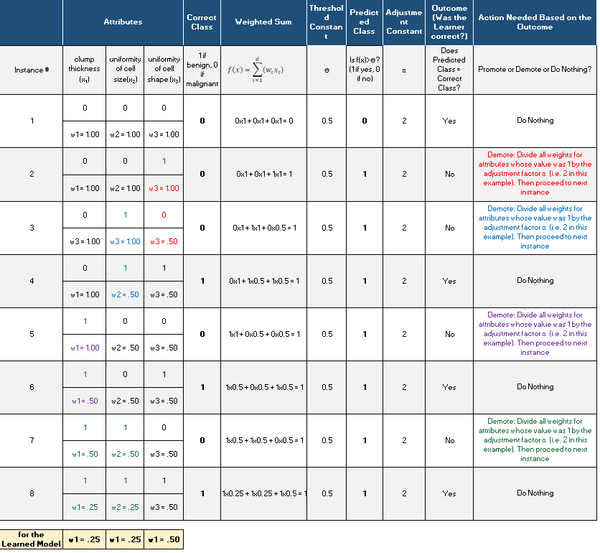
Here is what we have after we run Winnow2. We proceeded row-by-row in the original data set (one instance at a time), which generated this table:
Winnow2 Implementation
I implemented Winnow2 from scratch in Python and then ran the algorithm on a real-world data set, the breast cancer data set from the UCI Machine Learning Repository.
Preprocessing the Example Data Set – Breast Cancer
The breast cancer data set contains 699 instances, 10 attributes, and a class – malignant or benign. I transformed the attributes and classes into binary numbers (making them discrete is fine too, but they cannot be continuous) so that the algorithms could process the data properly and efficiently. If attribute value was greater than 5, the value was changed to 1, otherwise it was 0. I also changed Benign from 2 to 0 and Malignant from 4 to 1.
There were 16 missing attribute values in the data set, each denoted with a “?”. I chose a random number between 1 and 10 (inclusive) to fill in the data.
Finally, for each run, data sets were split into 67% for training and 33% for testing. Summary statistics were then calculated.
Here is the format the input data needs to take in the raw text/csv file. Attributes can be numerical or text, but in this example the were all numerical:
Columns (0 through N)
- 0: Instance ID
- 1: Attribute 1 (in binary)
- 2: Attribute 2 (in binary)
- 3: Attribute 3 (in binary)
- …
- N: Actual Class (in binary)
The program then adds 8 additional columns:
- N + 1: Weighted Sum (of the attributes)
- N + 2: Predicted Class (in binary)…Weighted Sum > 0? (1 if yes; 0 if no)
- N + 3: True Positive (1 if yes; O if no)
- N + 4: False Positive (1 if yes; 0 if no)
- N + 5: False Negative (1 if yes; 0 if no)
- N + 6: True Negative (1 if yes; 0 if no)
- N + 7: Promote (1 if yes; 0 if no) [for training set only]
- N + 8: Demote (1 if yes; 0 if no) [for training set only]
Here is a link to the input file after all that preprocessing was performed.
Here is a link to the output file after the algorithm below was run on the input file.
Here is a link to the final weights after the algorithm below was run on the input file.
Winnow2 Algorithm in Python, Coded From Scratch
Here is the code. Don’t be scared at how long the code is. I included a bunch of comments so that you know what is going on at each step. I recommend you copy and paste all this code into your favorite IDE. If there is a term or a piece of code that you have never seen before, look it up on Google (even the pros who have been at this a long time have to look things up on Google all the time! This is how you learn).
import pandas as pd # Import Pandas library
import numpy as np # Import Numpy library
# File name: winnow2.py
# Author: Addison Sears-Collins
# Date created: 5/31/2019
# Python version: 3.7
# Description: Implementation of the Winnow2 machine learning
# algorithm invented by Nick Littlestone. Used for 2-class classificaiton
# problems (e.g. cancer/no cancer....spam/not spam, etc...)
# Nick Littlestone (1988). "Learning Quickly When Irrelevant Attributes
# Abound: A New Linear-threshold Algorithm", Machine Learning 285–318(2)
# Required Data Set Format:
# Columns (0 through N)
# 0: Instance ID
# 1: Attribute 1 (in binary)
# 2: Attribute 2 (in binary)
# 3: Attribute 3 (in binary)
# ...
# N: Actual Class (in binary)
# This program then adds 8 additional columns:
# N + 1: Weighted Sum (of the attributes)
# N + 2: Predicted Class (in binary)...Weighted Sum > 0? (1 if yes; 0 if no)
# N + 3: True Positive (1 if yes; O if no)
# N + 4: False Positive (1 if yes; 0 if no)
# N + 5: False Negative (1 if yes; 0 if no)
# N + 6: True Negative (1 if yes; 0 if no)
# N + 7: Promote (1 if yes; 0 if no) [for training set only]
# N + 8: Demote (1 if yes; 0 if no) [for training set only]
################ INPUT YOUR OWN VALUES IN THIS SECTION ######################
ALGORITHM_NAME = "Winnow2"
THETA = 0.5 # This is the threshold constant for the Winnow2 algorithm
ALPHA = 2.0 # This is the adjustment constant for promotion & demotion
DATA_PATH = "breast_cancer_dataset.txt" # Directory where data set is located
TRAIN_WEIGHTS_FILE = "breast_cancer_winnow2_train_weights.txt" # Weights of learned model
TRAIN_OUT_FILE = "breast_cancer_winnow2_train_out.txt" # Training phase of the model
TEST_STATS_FILE = "breast_cancer_winnow2_test_stats.txt" # Testing statistics
TEST_OUT_FILE = "breast_cancer_winnow2_test_out.txt" # Testing phase of the model
SEPARATOR = "," # Separator for the data set (e.g. "\t" for tab data)
CLASS_IF_ONE = "Malignant" # If Class value is 1 (e.g. Malignant, Spam, etc.)
CLASS_IF_ZERO = "Benign" # If Class value is 0 (e.g. Benign, Not Spam, etc.)
TRAINING_DATA_PRCT = 0.67 # % of data set used for training
testing_data_prct = 1 - TRAINING_DATA_PRCT # % of data set used for testing
SEED = 99 # SEED for the random number generator. Default: 99
#############################################################################
# Read a text file and store records in a Pandas dataframe
pd_data = pd.read_csv(DATA_PATH, sep=SEPARATOR)
# Create a training dataframe by sampling random instances from original data.
# random_state guarantees that the pseudo-random number generator generates
# the same sequence of random numbers each time.
pd_training_data = pd_data.sample(frac=TRAINING_DATA_PRCT, random_state=SEED)
# Create a testing dataframe. Dropping the training data from the original
# dataframe ensures training and testing dataframes have different instances
pd_testing_data = pd_data.drop(pd_training_data.index)
# Convert training dataframes to Numpy arrays
np_training_data = pd_training_data.values
np_testing_data = pd_testing_data.values
#np_training_data = pd_data.values # Used for testing only
#np_testing_data = pd_data.values # Used for testing only
################ Begin Training Phase #####################################
# Calculate the number of instances, columns, and attributes in the data set
# Assumes 1 column for the instance ID and 1 column for the class
# Record the index of the column that contains the actual class
no_of_instances = np_training_data.shape[0]
no_of_columns = np_training_data.shape[1]
no_of_attributes = no_of_columns - 2
actual_class_column = no_of_columns - 1
# Initialize the weight vector. Initialize all weights to 1.
# First column of weight vector is not used (i.e. Instance ID)
weights = np.ones(no_of_attributes + 1)
# Create a new array that has 8 columns, initialized to 99 for each value
extra_columns_train = np.full((no_of_instances, 8),99)
# Add extra columns to the training data set
np_training_data = np.append(np_training_data, extra_columns_train, axis=1)
# Make sure it is an array of floats
np_training_data = np_training_data.astype(float)
# Build the learning model one instance at a time
for row in range(0, no_of_instances):
# Set the weighted sum to 0
weighted_sum = 0
# Calculate the weighted sum of the attributes
for col in range(1, no_of_attributes + 1):
weighted_sum += (weights[col] * np_training_data[row,col])
# Record the weighted sum into column N + 1, the column just to the right
# of the actual class column
np_training_data[row, actual_class_column + 1] = weighted_sum
# Set the predicted class to 99
predicted_class = 99
# Learner's prediction: Is the weighted sum > THETA?
if weighted_sum > THETA:
predicted_class = 1
else:
predicted_class = 0
# Record the predicted class into column N + 2
np_training_data[row, actual_class_column + 2] = predicted_class
# Record the actual class into a variable
actual_class = np_training_data[row, actual_class_column]
# Initialize the prediction outcomes
# These variables are standard inputs into a "Confusion Matrix"
true_positive = 0 # Predicted class = 1; Actual class = 1 (hit)
false_positive = 0 # Predicted class = 1; Actual class = 0 (false alarm)
false_negative = 0 # Predicted class = 0; Actual class = 1 (miss)
true_negative = 0 # Predicted class = 0; Actual class = 0
# Determine the outcome of the Learner's prediction
if predicted_class == 1 and actual_class == 1:
true_positive = 1
elif predicted_class == 1 and actual_class == 0:
false_positive = 1
elif predicted_class == 0 and actual_class == 1:
false_negative = 1
else:
true_negative = 1
# Record the outcome of the Learner's prediction
np_training_data[row, actual_class_column + 3] = true_positive
np_training_data[row, actual_class_column + 4] = false_positive
np_training_data[row, actual_class_column + 5] = false_negative
np_training_data[row, actual_class_column + 6] = true_negative
# Set the promote and demote variables to 0
promote = 0
demote = 0
# Promote if false negative
if false_negative == 1:
promote = 1
# Demote if false positive
if false_positive == 1:
demote = 1
# Record if either a promotion or demotion is needed
np_training_data[row, actual_class_column + 7] = promote
np_training_data[row, actual_class_column + 8] = demote
# Run through each attribute and see if it is equal to 1
# If attribute is 1, we need to either demote or promote (adjust the
# corresponding weight by ALPHA).
if demote == 1:
for col in range(1, no_of_attributes + 1):
if(np_training_data[row,col] == 1):
weights[col] /= ALPHA
if promote == 1:
for col in range(1, no_of_attributes + 1):
if(np_training_data[row,col] == 1):
weights[col] *= ALPHA
# Open a new file to save the weights
outfile1 = open(TRAIN_WEIGHTS_FILE,"w")
# Write the weights of the Learned model to a file
outfile1.write("----------------------------------------------------------\n")
outfile1.write(" " + ALGORITHM_NAME + " Training Weights\n")
outfile1.write("----------------------------------------------------------\n")
outfile1.write("Data Set : " + DATA_PATH + "\n")
outfile1.write("\n----------------------------\n")
outfile1.write("Weights of the Learned Model\n")
outfile1.write("----------------------------\n")
for col in range(1, no_of_attributes + 1):
colname = pd_training_data.columns[col]
s = str(weights[col])
outfile1.write(colname + " : " + s + "\n")
# Write the relevant constants used in the model to a file
outfile1.write("\n")
outfile1.write("\n")
outfile1.write("-----------\n")
outfile1.write("Constants\n")
outfile1.write("-----------\n")
s = str(THETA)
outfile1.write("THETA = " + s + "\n")
s = str(ALPHA)
outfile1.write("ALPHA = " + s + "\n")
# Close the weights file
outfile1.close()
# Print the weights of the Learned model
print("----------------------------------------------------------")
print(" " + ALGORITHM_NAME + " Results")
print("----------------------------------------------------------")
print("Data Set : " + DATA_PATH)
print()
print()
print("----------------------------")
print("Weights of the Learned Model")
print("----------------------------")
for col in range(1, no_of_attributes + 1):
colname = pd_training_data.columns[col]
s = str(weights[col])
print(colname + " : " + s)
# Print the relevant constants used in the model
print()
print()
print("-----------")
print("Constants")
print("-----------")
s = str(THETA)
print("THETA = " + s)
s = str(ALPHA)
print("ALPHA = " + s)
print()
# Print the learned model runs in binary form
print("-------------------------------------------------------")
print("Learned Model Runs of the Training Data Set (in binary)")
print("-------------------------------------------------------")
print(np_training_data)
print()
print()
# Convert Numpy array to a dataframe
df = pd.DataFrame(data=np_training_data)
# Replace 0s and 1s in the attribute columns with False and True
for col in range(1, no_of_attributes + 1):
df[[col]] = df[[col]].replace([0,1],["False","True"])
# Replace values in Actual Class column with more descriptive values
df[[actual_class_column]] = df[[actual_class_column]].replace([0,1],[CLASS_IF_ZERO,CLASS_IF_ONE])
# Replace values in Predicted Class column with more descriptive values
df[[actual_class_column + 2]] = df[[actual_class_column + 2]].replace([0,1],[CLASS_IF_ZERO,CLASS_IF_ONE])
# Change prediction outcomes to more descriptive values
for col in range(actual_class_column + 3,actual_class_column + 9):
df[[col]] = df[[col]].replace([0,1],["No","Yes"])
# Rename the columns
df.rename(columns={actual_class_column + 1 : "Weighted Sum" }, inplace = True)
df.rename(columns={actual_class_column + 2 : "Predicted Class" }, inplace = True)
df.rename(columns={actual_class_column + 3 : "True Positive" }, inplace = True)
df.rename(columns={actual_class_column + 4 : "False Positive" }, inplace = True)
df.rename(columns={actual_class_column + 5 : "False Negative" }, inplace = True)
df.rename(columns={actual_class_column + 6 : "True Negative" }, inplace = True)
df.rename(columns={actual_class_column + 7 : "Promote" }, inplace = True)
df.rename(columns={actual_class_column + 8 : "Demote" }, inplace = True)
# Change remaining columns names from position numbers to descriptive names
for pos in range(0,actual_class_column + 1):
df.rename(columns={pos : pd_data.columns[pos] }, inplace = True)
print("-------------------------------------------------------")
print("Learned Model Runs of the Training Data Set (readable) ")
print("-------------------------------------------------------")
# Print the revamped dataframe
print(df)
# Write revamped dataframe to a file
df.to_csv(TRAIN_OUT_FILE, sep=",", header=True)
################ End Training Phase #####################################
################ Begin Testing Phase ######################################
# Calculate the number of instances, columns, and attributes in the data set
# Assumes 1 column for the instance ID and 1 column for the class
# Record the index of the column that contains the actual class
no_of_instances = np_testing_data.shape[0]
no_of_columns = np_testing_data.shape[1]
no_of_attributes = no_of_columns - 2
actual_class_column = no_of_columns - 1
# Create a new array that has 6 columns, initialized to 99 for each value
extra_columns_test = np.full((no_of_instances, 6),99)
# Add extra columns to the testing data set
np_testing_data = np.append(np_testing_data, extra_columns_test, axis=1)
# Make sure it is an array of floats
np_testing_data = np_testing_data.astype(float)
# Test the learning model one instance at a time
for row in range(0, no_of_instances):
# Set the weighted sum to 0
weighted_sum = 0
# Calculate the weighted sum of the attributes
for col in range(1, no_of_attributes + 1):
weighted_sum += (weights[col] * np_testing_data[row,col])
# Record the weighted sum into column N + 1, the column just to the right
# of the actual class column
np_testing_data[row, actual_class_column + 1] = weighted_sum
# Set the predicted class to 99
predicted_class = 99
# Learner's prediction: Is the weighted sum > THETA?
if weighted_sum > THETA:
predicted_class = 1
else:
predicted_class = 0
# Record the predicted class into column N + 2
np_testing_data[row, actual_class_column + 2] = predicted_class
# Record the actual class into a variable
actual_class = np_testing_data[row, actual_class_column]
# Initialize the prediction outcomes
# These variables are standard inputs into a "Confusion Matrix"
true_positive = 0 # Predicted class = 1; Actual class = 1 (hit)
false_positive = 0 # Predicted class = 1; Actual class = 0 (false alarm)
false_negative = 0 # Predicted class = 0; Actual class = 1 (miss)
true_negative = 0 # Predicted class = 0; Actual class = 0
# Determine the outcome of the Learner's prediction
if predicted_class == 1 and actual_class == 1:
true_positive = 1
elif predicted_class == 1 and actual_class == 0:
false_positive = 1
elif predicted_class == 0 and actual_class == 1:
false_negative = 1
else:
true_negative = 1
# Record the outcome of the Learner's prediction
np_testing_data[row, actual_class_column + 3] = true_positive
np_testing_data[row, actual_class_column + 4] = false_positive
np_testing_data[row, actual_class_column + 5] = false_negative
np_testing_data[row, actual_class_column + 6] = true_negative
# Convert Numpy array to a dataframe
df = pd.DataFrame(data=np_testing_data)
# Replace 0s and 1s in the attribute columns with False and True
for col in range(1, no_of_attributes + 1):
df[[col]] = df[[col]].replace([0,1],["False","True"])
# Replace values in Actual Class column with more descriptive values
df[[actual_class_column]] = df[[actual_class_column]].replace([0,1],[CLASS_IF_ZERO,CLASS_IF_ONE])
# Replace values in Predicted Class column with more descriptive values
df[[actual_class_column + 2]] = df[[actual_class_column + 2]].replace([0,1],[CLASS_IF_ZERO,CLASS_IF_ONE])
# Change prediction outcomes to more descriptive values
for col in range(actual_class_column + 3,actual_class_column + 7):
df[[col]] = df[[col]].replace([0,1],["No","Yes"])
# Rename the columns
df.rename(columns={actual_class_column + 1 : "Weighted Sum" }, inplace = True)
df.rename(columns={actual_class_column + 2 : "Predicted Class" }, inplace = True)
df.rename(columns={actual_class_column + 3 : "True Positive" }, inplace = True)
df.rename(columns={actual_class_column + 4 : "False Positive" }, inplace = True)
df.rename(columns={actual_class_column + 5 : "False Negative" }, inplace = True)
df.rename(columns={actual_class_column + 6 : "True Negative" }, inplace = True)
df_numerical = pd.DataFrame(data=np_testing_data) # Keep the values in this dataframe numerical
df_numerical.rename(columns={actual_class_column + 3 : "True Positive" }, inplace = True)
df_numerical.rename(columns={actual_class_column + 4 : "False Positive" }, inplace = True)
df_numerical.rename(columns={actual_class_column + 5 : "False Negative" }, inplace = True)
df_numerical.rename(columns={actual_class_column + 6 : "True Negative" }, inplace = True)
# Change remaining columns names from position numbers to descriptive names
for pos in range(0,actual_class_column + 1):
df.rename(columns={pos : pd_data.columns[pos] }, inplace = True)
print("-------------------------------------------------------")
print("Learned Model Predictions on Testing Data Set")
print("-------------------------------------------------------")
# Print the revamped dataframe
print(df)
# Write revamped dataframe to a file
df.to_csv(TEST_OUT_FILE, sep=",", header=True)
# Open a new file to save the summary statistics
outfile2 = open(TEST_STATS_FILE,"w")
# Write to a file
outfile2.write("----------------------------------------------------------\n")
outfile2.write(ALGORITHM_NAME + " Summary Statistics (Testing)\n")
outfile2.write("----------------------------------------------------------\n")
outfile2.write("Data Set : " + DATA_PATH + "\n")
# Write the relevant stats to a file
outfile2.write("\n")
outfile2.write("Number of Test Instances : " +
str(np_testing_data.shape[0])+ "\n")
tp = df_numerical["True Positive"].sum()
s = str(int(tp))
outfile2.write("True Positives : " + s + "\n")
fp = df_numerical["False Positive"].sum()
s = str(int(fp))
outfile2.write("False Positives : " + s + "\n")
fn = df_numerical["False Negative"].sum()
s = str(int(fn))
outfile2.write("False Negatives : " + s + "\n")
tn = df_numerical["True Negative"].sum()
s = str(int(tn))
outfile2.write("True Negatives : " + s + "\n")
accuracy = (tp + tn)/(tp + tn + fp + fn)
accuracy *= 100
s = str(accuracy)
outfile2.write("Accuracy : " + s + "%\n")
specificity = (tn)/(tn + fp)
specificity *= 100
s = str(specificity)
outfile2.write("Specificity : " + s + "%\n")
precision = (tp)/(tp + fp)
precision *= 100
s = str(precision)
outfile2.write("Precision : " + s + "%\n")
recall = (tp)/(tp + fn)
recall *= 100
s = str(recall)
outfile2.write("Recall : " + s + "%\n")
neg_pred_value = (tn)/(tn + fn)
neg_pred_value *= 100
s = str(neg_pred_value)
outfile2.write("Negative Predictive Value : " + s + "%\n")
miss_rate = (fn)/(fn + tp)
miss_rate *= 100
s = str(miss_rate)
outfile2.write("Miss Rate : " + s + "%\n")
fall_out = (fp)/(fp + tn)
fall_out *= 100
s = str(fall_out)
outfile2.write("Fall-Out : " + s + "%\n")
false_discovery_rate = (fp)/(fp + tp)
false_discovery_rate *= 100
s = str(false_discovery_rate)
outfile2.write("False Discovery Rate : " + s + "%\n")
false_omission_rate = (fn)/(fn + tn)
false_omission_rate *= 100
s = str(false_omission_rate)
outfile2.write("False Omission Rate : " + s + "%\n")
f1_score = (2 * tp)/((2 * tp) + fp + fn)
s = str(f1_score)
outfile2.write("F1 Score: " + s)
# Close the weights file
outfile2.close()
# Print statistics to console
print()
print()
print("-------------------------------------------------------")
print(ALGORITHM_NAME + " Summary Statistics (Testing)")
print("-------------------------------------------------------")
print("Data Set : " + DATA_PATH)
# Print the relevant stats to the console
print()
print("Number of Test Instances : " +
str(np_testing_data.shape[0]))
s = str(int(tp))
print("True Positives : " + s)
s = str(int(fp))
print("False Positives : " + s)
s = str(int(fn))
print("False Negatives : " + s)
s = str(int(tn))
print("True Negatives : " + s)
s = str(accuracy)
print("Accuracy : " + s + "%")
s = str(specificity)
print("Specificity : " + s + "%")
s = str(precision)
print("Precision : " + s + "%")
s = str(recall)
print("Recall : " + s + "%")
s = str(neg_pred_value)
print("Negative Predictive Value : " + s + "%")
s = str(miss_rate)
print("Miss Rate : " + s + "%")
s = str(fall_out)
print("Fall-Out : " + s + "%")
s = str(false_discovery_rate)
print("False Discovery Rate : " + s + "%")
s = str(false_omission_rate)
print("False Omission Rate : " + s + "%")
s = str(f1_score)
print("F1 Score: " + s)
###################### End Testing Phase ######################################
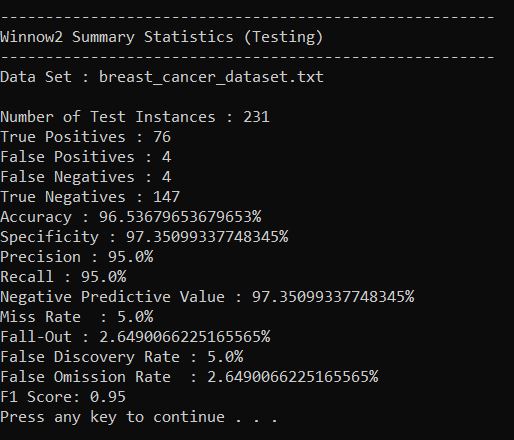
Output Statistics of Winnow2 on the Breast Cancer Data Set
Here is a link to a screenshot of the summary statistics: